Interview
Antibiotics from the bacterial world
Experts are convinced that there are undetected antibiotic substances awaiting discovery. They are getting ready to find them.
Considering that the challenge of multi-resistant bacteria has been known for years and that, according to estimates by the Robert Koch Institute, it claims the lives of 10,000 to 20,000 people every year in Germany alone, it might seem surprising that, in the last 15 years, little effort of large pharmaceutical companies went into researching new antibiotics. The Center for Systems-Based Antibiotics Research, short Cesar, is currently being established to provide an infrastructure that is unique in Germany that will aid the search for new, antibiotic substances. Professor Julia Bandow, Chair of Applied Microbiology at RUB, coordinates the Research Center, which is being set up in collaboration with the Lead Discovery Center, Dortmund. In this interview she outlines the approach.
Professor Bandow, why has antibiotics research been neglected for so long?
Politicians long thought that antibiotic research and development more or less exclusively was the domain of the pharmaceutical industry, even when major pharmaceutical companies had exited this area. Unfortunately, the industry as a whole had very much bet on the wrong horse: for a decade, they had been searching for compounds that specifically target individual, vital proteins of pathogens, and they indeed found them. However, it turned out that when only one single target is addressed, spontaneous mutations in the relevant gene can quickly cause resistance. So far, none of these efforts have led to new drugs being launched in the market.

Another problem was that, although many substances interfere with the function of bacterial proteins in the test tube, they are either unable to enter the cells, or the cells transport them back out using molecular pumps. As a result, the industry has been left with primarily introducing new derivatives of known antibiotics to the market for a long time. Tragically, this applies to the industry worldwide.
In what way do you pursue a different approach with Cesar?
We are focusing on two aspects: on the one hand, we want to find new substances, i.e. new classes of substances to replace the existing ones, which are becoming less and less effective. To this end, we have to find them and produce them in sufficient quantities to be able to test them and, if necessary, develop derivatives.
The other aspect is the analysis of the mechanisms of action. We want to know: what do these substances do? Are they effective only against bacteria or do they also affect components of human cells?
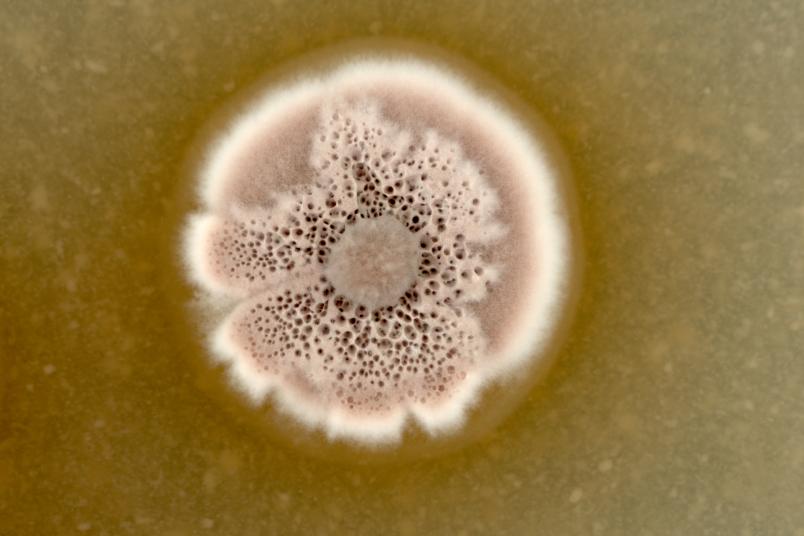
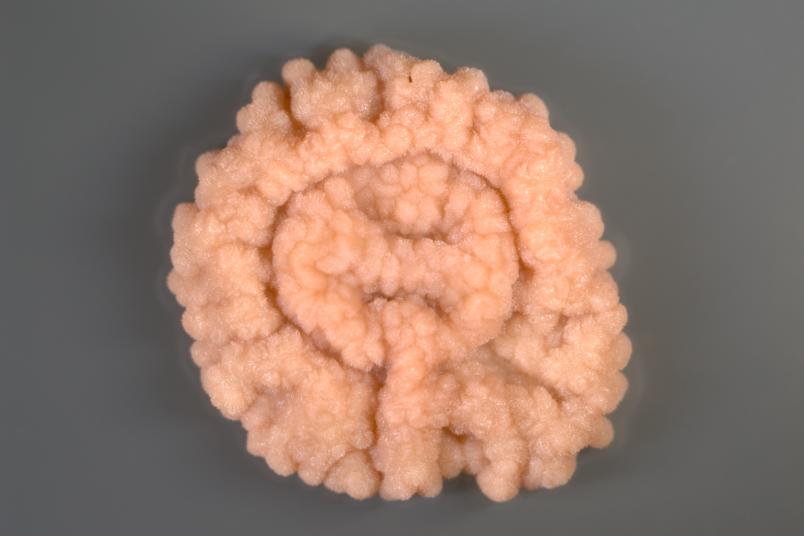
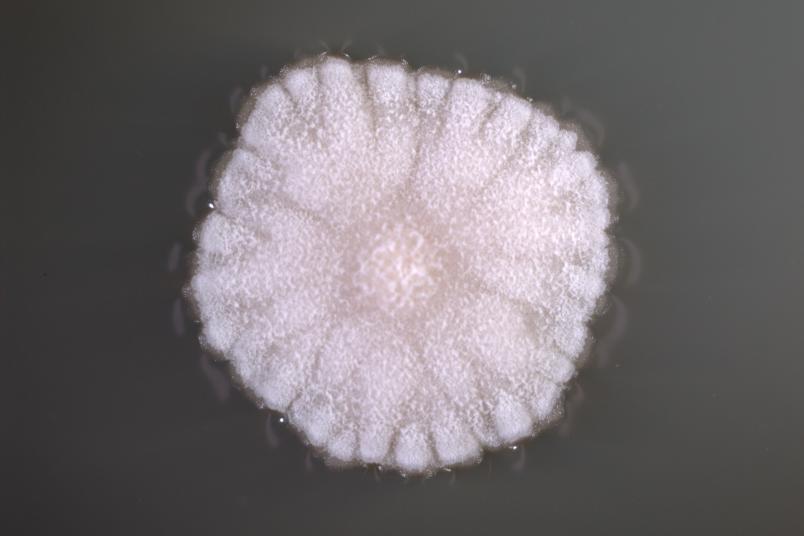
We use global methods; that means we examine all metabolic products that microorganisms secrete and all proteins that they synthesise. Many microorganisms that compete with others for resources in their habitats release substances that inhibit competitors, i.e. substances that have an antibiotic effect.
We use streptomyces as source organisms. Streptomyces are bacteria that live in the soil and are particularly good chemists, as they release many substances. Almost all natural substances currently used as antibiotics are produced either by this group of bacteria or by fungi. Since detection methods have improved considerably in recent years, we hope to uncover yet unknown substances. Quite promising groundwork was recently laid: in a study from 2018, Christoph Senges from my research group detected over 1,000 substances that can be produced by a single streptomyces strain.
How do you handle such a huge number of substances?
Let’s take Streptomyces chartreusis, for example, which harbours approximately 128 gene clusters for the synthesis of natural substances. This diversity is difficult to tackle using comparative genetic analyses, because where should we start? The products of most biosynthetic gene clusters can’t easily be predicted.
In Cesar, we take a different approach: we look at the secreted metabolic products and cluster structurally related ones. The aim is to then purify representative substances for the clusters in order to elucidate their structure and test them for antibiotic activity. Based on the representatives, we can subsequently annotate the corresponding cluster of substances.
Much is still speculation; for example, is the size of a cluster, i.e. the number of variants of a substance an indication that this group of compoundss is of particular importance for the producer?
In the first step, however, the team will focus on setting up the infrastructure.
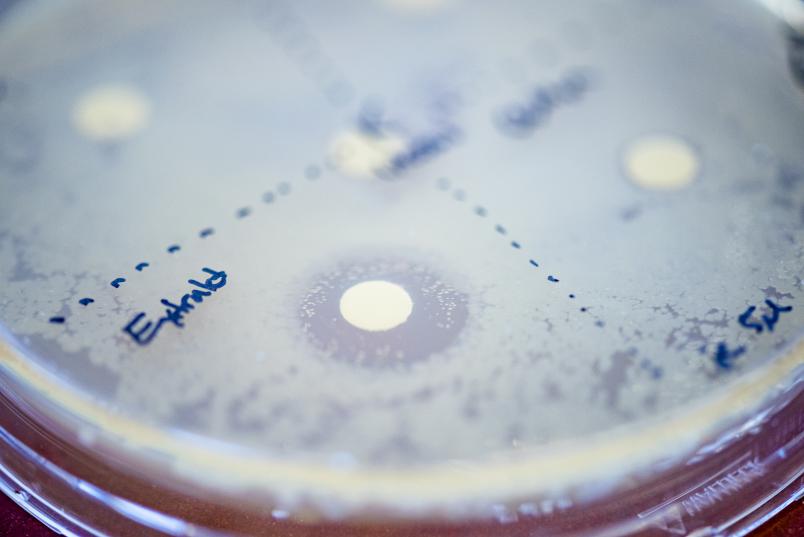
What does this entail?
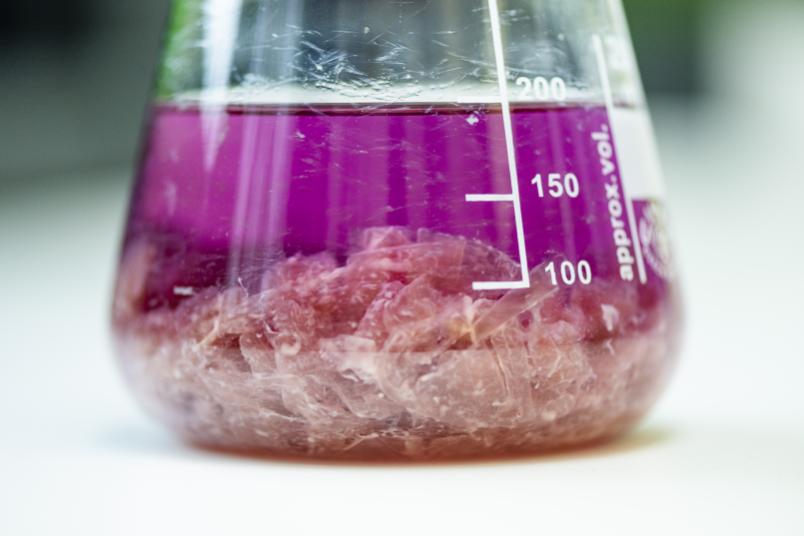
We’re obtaining equipment for analysing the metabolome of micro-organisms, i.e. all the products that are secreted, and for analysing the proteome, i.e. all the proteins they produce. We also need other equipment for compound purification, such as shakers, centrifuges, and chromatography systems. The equipment must be integrated into workflows that serve to complete the various experimental stages. The infrastructure will form the basis for research and development projects that are going to focus on the discovery and characterisation of new substances.

Another aim is to build expertise in natural product purification: experience shows that 80 per cent of the undesirable substances can be removed quite easily, but the remaining 20 per cent are more persistent. Here, several methods are required that exploit different properties of substances in order to separate them.
Since by training most of us are biologists, we are very well-versed in proteomics and the analysis of mechanisms of action; now, we look forward to expanding our expertise in chemistry, to meet the different challenges the substance pose.
Are you optimistic that new antibiotics will be discovered?
I’m sure that there are novel groups of substances with antibiotic effects to be discovered and that we can find them.